The latest data mining scheme sort out!
Author:Data School Thu Time:2022.07.05

There are 1500 words in this article, it is recommended to read for 9 minutes
This article takes you immersive to experience the participation process from 0 to 1 in the prediction of diabetic genetic risk prediction.
Introduction to the topic
HKUST Xunfei: Diabetes Genetic Risk Test Challenge. Background: As of 2022, there are nearly 130 million diabetic patients in China. Causes of Chinese diabetes are affected by various factors such as lifestyle, aging, urbanization, and family genetic. At the same time, diabetic patients tend to be younger.
Diabetes can cause complications of cardiovascular, kidney, and cerebrovascular complications. Therefore, it has very important clinical significance to accurately diagnose individuals with diabetes. Early genetic risk prediction of diabetes will help prevent the occurrence of diabetes.
The address of the event:
http://challenge.xfyun.cn/topic/info?Type=diabetesch=ds22-dw-gzy01
Source code:
https://github.com/datawhalechina/competition-baseline
Tie task
In this competition, you need to build a diabetic genetic risk prediction model through training data sets, and then predict whether the individual of the test data set has diabetes, and help diabetic patients to solve this "sweet trouble" with us.
Tournament data
The topic data is composed of training sets and test collection. The specific situation is as follows:
Training set: There are 5070 pieces of data, which is used to build your prediction model
Test set: There are 1,000 pieces of data for verifying the performance of the predicted model.
Among them, the training collection data includes 9 fields: gender, year of birth, weight index, family history of diabetes, diastolic pressure, oral sugar resistance test, insulin release experiments, triceis muscle fold thickness, diabetes signs (data labels) Essence
Grading
The F1-SCORE indicator in the binary class task is evaluated. The larger the F1-SCORE shows that the better the predicted model, the better the F1-SCORE definition is as follows:
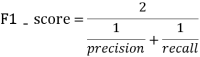
in:
Treatment Baseline import data
Import Pandas as PD Import LightGBM
Pre -processing
data1 = pd.read_csv ('' competition training set .csv ', encoding =' gbk ') data2 = pd.read_csv (' 'competition test set .csv', enCoding = 'gbk') #Label marked as -1 data2 ['Diabetes logo'] =-1 #<<< data = pd.concat ([data1, data2], axis = 0, iGnore_index = true) characteristics "" "" "" "" "" "" "" " The normal value of the human body weight index of the human body is between 18.5-24 below 18.5 is too light weight index between 24-27 is Overweight 27 or above is considered obesity higher than 32 is very obese.
not > Elif 18.5 <= A <= 24: Return 1 Elif 24 CODE> "" " Return 2 Elif 27 Return 3 Else: Return 4 Data ['BMI'] = Data ['Weight Index'] .apply (BMI) Data ['Birth Year'] = 2022-Data ['Birth Year'] #Stead to age #Diabetes Family History "" " No record Uncle or aunt has one with diabetes/uncle or aunt with diabetes with diabetes One of the parents suffering from diabetes "" " defff (A): if A == 'No record': Return 0 Elif A == 'Uncle or Auntie has a diabetes' or a ==' Uncle or Auntie. Diabetes ': Return 1 Else: Return 2 Data [' Diabetes Family History '] = Data [' Diabetes Family History '] .apply (FHOD) Data [' diastolic pressure '] = data [' diastolic pressure '] .Fillna (-1) The range of diastolic blood pressure is 60-90 " "" Def Dbp (a): IF a <60: Return 0 Elif 60 <= A Return 1 Elif A & 90: Return 2 Else: Return a data ['dbp' ] = Data ['Solid Pressure'] .apply (DBP) Data Training Data/Test Data Preparation
Train = Data [Data ['Diabetes logo']! =-1] test = data [data ['Diabetes logo'] ==-1] Train_label = TRAIN ['With diabetes logo'] Train = train.drop (['number', 'with diabetes logo'], axis = 1) > test = test.drop (['number', 'with diabetes logo'], axis = 1)
Build a model
def select_by_lgb(train_data,train_label,test_data,random_state=2022,metric='auc',num_round=300): clf=lightgbm train_matrix=clf.Dataset( Train_data, label = train_label) Params = 'Boosting_type': 'gbdt', ' Learning_raate ': 0.1, ' metric ': metric, ' seed ': 2020, ' nthread ': -1 Model = CLF.Train (Params, Train_Matrix, NUM_ROUND) Pre_y = Model.predict (TEST_DATA) Return Pre_y Model verification
Test_data = Select_by_LGB (Train, Train_label, Test) Pre_y = pdataframe (test_data) pre_y ['label'] = pre_y [0] .apply (lambdada. x: 1 if x & 0.5 else 0) Result = pd.read_csv ('Submit example .csv') Result ['label'] = pre_y ['label'] Result.to_csv ('Baseline.csv', Index = False)
Proposal
The model part of this question uses a fairly simple method. The LightGBM algorithm does not verify the cross -data data, and only uses the default parameter. Therefore, the players can further optimize the parameter optimization and the model fusion on the basis of this Baseline. The constructing feature engineering construction in this open algorithm has been demonstrated. Players can further dig the characteristics according to the medical characteristics of diabetes, thereby improving the results of the competition ~


- END -
Good medicine daily science 丨 to vaccination during the high incidence of influenza is the most effective preventive means

Influenza is one of the severe respiratory infectious diseases. Recently, the nati...
Zhejiang Provincial Prevention and Control Office: Adjust the health management measures of key personnel

On July 1, the Office of the Leading Group of the New Coronatte Pneumonic Pneumoni...