Cattle!In the past few months, for a few seconds now
Author:Journal of China Science Time:2022.09.20
Text | Li Muzi
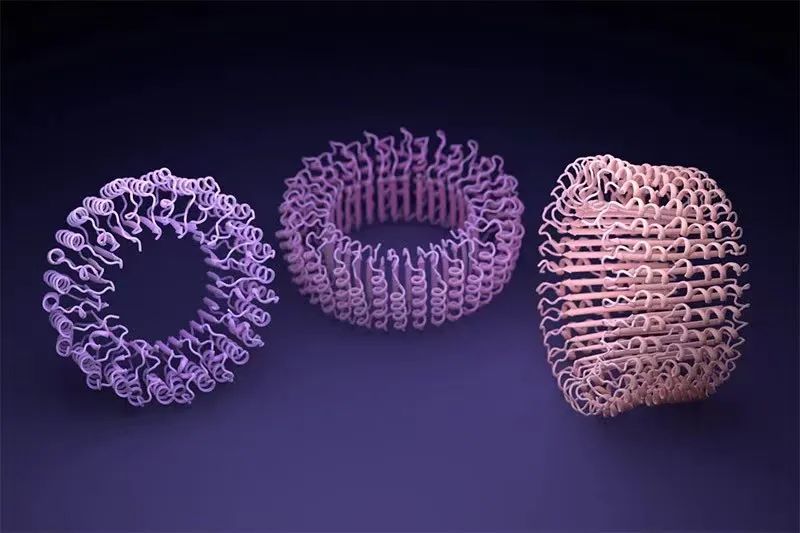
Artificial intelligence tools are helping scientists design protein that is different from any natural structure. Image source: Ian C Haydon/UW Institute for Protein Design
In June this year, the South Korean regulatory agency approved the first new protein -based new protein -based new -type coronary pneumonia vaccine. The vaccine is based on a spherical protein "nano particles" and was developed by researchers 10 years ago through trial and error over labor type.
Now, with the huge progress of artificial intelligence (AI), a team led by the University of Seattle Washington (UW) biochemist David Baker in the United States only takes a few seconds -instead of a few months -it can be designed to design this kind of molecular. Related studies were published on September 15th.
These efforts are part of the scientific change. AI tools such as DeepMind's protein structure prediction software Alphafold are favored by life scientists. In July this year, DeepMind revealed that the latest version of Alphafold has predicted the structure of all protein. In recent months, AI tools have experienced explosive growth -some of them are based on Alphafold, which can quickly create new proteins. Before that, this was a difficult job with a high failure rate.
Baker's laboratory has been creating new protein in the past 30 years. His laboratory has developed a software called Rosetta in the 1990s, which can divide this process into multiple steps. Initially, researchers imagined the shape of a new protein -usually put together other protein fragments together, and then the software derives the amino acid sequence corresponding to the shape.
However, when making these "drafts" proteins in the laboratory, it is rarely folded into the required shapes. On the contrary, they are eventually stuck in different states. Therefore, another step is needed to adjust the protein sequence to make it only fold into a single required structure. Harvard University evolved biologist Sergey Ovchinnikov, who had worked at Baker laboratory, said that this step involves all methods that mimics different sequences may be folded, and the calculation cost is high. "It may take 10,000 computers to run for several weeks to complete this."
Ovchinnikov said, but by adjusting Alphafold and other AI programs, this time -consuming step can be completed instantly. In a method called "Phantom" developed by the Baker team, the researchers will predict the random amino acid sequence input structure prediction network; according to the network prediction, it changes its structure and makes it more like protein. In a papers in 2021, the Baker team created more than 100 small "hallucinations" protein in the laboratory, of which about 1/5 of the protein was similar to the predictive shape.
A similar tools developed by Alphafold and Baker laboratory RosettaFold are trained to predict the structure of a single protein chain. But researchers quickly discovered that this network can also simulate a combination of multi -interaction protein. On this basis, the Baker team believes that they can assemble the protein self -assembly into different shapes and sizes of nanoparticles, similar to the protein involved in the above -mentioned new coronary pneumonia vaccine. The Baker team firmly believes that creating a new protein in the laboratory is the final test of their methods.
Baker said that experiments are essential when designing proteins with specific tasks. In July, his team described an AI method that allowed researchers to embed new protein in specific sequences or structures. They use these methods to design enzymes that catalyzes specific reactions, proteins that can be combined with other molecules, and a protein that can be used to combat respiratory virus vaccines, and respiratory viruses are the main causes of infant hospitalization.
Last year, DeepMind established a company called "Type Laboratory" in London, England, and intends to apply ALPhafold and other AI tools to drug discovery. DeepMind CEO Demis Hassabis said protein design is an obvious and promising application of deep learning technology, especially Alphafold. "Although we have done a lot of work in the field of protein design, it is just the beginning."
Related thesis information:
https://doi.org/10.1038/d41586-02947-7
"China Science News" (2022-09-20 The 2nd edition of the original title "Artificial Intelligence Design" Original "New Protein")
Edit | Zhao Lu
Capture | Zhihai

- END -
Henan Province's polymer materials and radiation processing industry technology innovation strategy alliance was established

Dahe Daily · Yu Video Reporter GaojuOn July 31, Henan Province's polymer material...
The main channel of the digital economy ignites the new engine of high -quality development

Ten years of hard work, ten years of wonderful butterfly changed. The mobile netwo...